Site Links:
|
Descent of Man In this lecture, beginners can familiarize themselves with basic information and terms used to describe the evolution of humanity beginning with the origin of primates through the comings and goings of Genus Homo. |

Cloned animals don't look alike
Posted: Saturday, December 28, 2002
Author: Tina Hesman Post-Dispatch
Filed: 12/28/2002, 10:36:21 PM
Source: St. Louis Post-Dispatch
Cloned animals may be exact copies genetically, but they don't look alike
When scientists at Clonaid announced Friday that they have successfully cloned a human being, many researchers expressed skepticism. But even if the baby girl is a genetic replica of her mother, the child is likely to look different from the woman who spawned her and behave differently, experts say. And the human clone could be at risk for a myriad of health problems such as those seen in cloned animals.
Scientists must make multiple attempts to produce clones. For every clone produced, many more die in the test-tube or during gestation, and scientists don't yet understand why or what to do about it.
"We have ideas about what the root causes are, but we don't have any idea about how to fix it," said Randall Prather, a reproductive biologist at the University of Missouri in Columbia. Prather has cloned pigs in a project that aims at producing organs for transplant into humans.
Although the pigs are genetic carbon copies of one another, the animals have differences in appearance, Prather said. Some pigs are taller than others or have different markings, he said.
Those differences are probably not due to changes in the animals' DNA, Prather said. He attributes the differences among his pigs and other cloned animals to imprinting - modifications to DNA that affect how genes are turned on and off.
Every cell in an animal's (or person's) body contains a complete set of genes for making the organism. But just as parents need instructions for assembling bicycles from a jumble of parts, so people and animals need directions for building bodies. Imprinting provides some of those directions.
That imprinting makes an impression that can last through all the manipulations of cloning. If imprinting is not fully erased, a cloned animal may have abnormalities.
Cloned mice often become obese in old age.
Dolly the sheep, the first animal cloned by nuclear transfer, is pudgy and arthritic.
Some cloned animals can't straighten their legs fully because their tendons contract.
Other cloned animals have weakened immune systems or enlarged hearts.
The most obvious evidence that clones aren't exact replicas of their genetic donors is CC, a cat cloned by scientists at Genetic Savings and Clone in College Station, Texas. The researchers took the genetic information used to make CC from a calico cat called Rainbow. The cats look completely different despite their identical genetic makeup. Rainbow's coat is splotched with orange and black patches, but CC is a black and white tiger tabby.
The researchers who created CC say that her mysterious lack of orange spots could be due to a failure to reset a type of imprinting known as X-linked inactivation.
Female cats, humans and other mammals have two X-chromosomes. Males carry only one X-chromosome and one Y-chromosome. To balance what would otherwise be a double dose of X genes, one of the X-chromosomes in each cell of a female animal's body is inactivated. The process seems to be completely random.
In humans, the effect is not usually noticeable, but in cats, X-inactivation affects a gene that controls coat color. One variant of the gene makes the fur orange, another version of the gene produces black fur. In calico cats, the X-chromosome containing the orange version of the gene gets shut off in some spots. That means the X-chromosome with the black gene is still active in those patches and produces splotches of black fur. In other spots, the X-chromosome containing the black gene is inactivated, producing orange patches.
Since CC has no orange fur, the researchers speculate that her genetic material was taken from a cell in which the X-chromosome carrying the orange gene was shut off. That leaves the year-old cat with only black patches, but indicates that CC could have other problems with her genetic programming.
Because humans also inactivate X-chromosomes, a cloned baby girl might have a similar problem. Experiments with cloned mice showed no defect in resetting inactive X-chromosomes, but the mice did have other differences in gene regulation.
![]() Send page by E-Mail
Send page by E-Mail
Posted: Thursday, December 26, 2002
Did Early Humans Mate With The Locals? Human Genome Data Cast Doubt On "Replacement Theory" Of Human Evolution
Source: University Of Utah
A new analysis of human genetic history deals a blow to the theory that early people moved out of Africa and completely replaced local populations elsewhere in the world. The findings suggest there was at least limited interbreeding between our African ancestors and the residents of areas where they settled.
"The new data seem to suggest that early human pioneers moving out of Africa starting 80,000 years ago did not completely replace local populations in the rest of the world," says Henry Harpending, a University of Utah anthropology professor and co-author of the new study. "There is instead some sign of interbreeding."
If that conclusion is correct, it contradicts the "replacement theory" of human evolution - a theory Harpending has advocated for more than a decade.
"Hypotheses are called into question by data every day in science. That's the way it works," he says.
The journal Proceedings of the National Academy of Sciences is publishing the new findings in its online edition the week of Dec. 23, 2002. The study's 20 co-authors include three from the University of Utah: Harpending; Alan Rogers, also a professor of anthropology; and Stephen Wooding, a postdoctoral researcher in human genetics.
The study was led by anthropologist Stephen Sherry and mathematician Gabor Marth of the National Center for Biotechnology Information at the National Institutes of Health in Bethesda, Md. Sherry is a former student of Harpending's when both were at Pennsylvania State University. Other co-authors of the new study are from the Washington University School of Medicine in St. Louis, The Johns Hopkins University School of Medicine in Baltimore and the University of California, San Francisco.
Most anthropologists agree human ancestors first spread out of Africa roughly 1.8 million years ago, establishing new populations in Europe, Asia and elsewhere. The "multiregional theory" holds modern humans evolved from those multiple populations. The competing "replacement theory" says that the local populations, including Europe's Neanderthals, went extinct when they were replaced roughly between 80,000 and 30,000 years ago by another wave of human immigrants from Africa.
Scientists can analyze ancient genetic mutations in modern people to learn about how humans evolved and the size of the human population over time. Mutations occur at a relatively steady rate over time. If the human population were large at a specific point in prehistoric time, more mutations would occur, resulting in greater diversity in genetic mutations found in modern people. A small population of human ancestors would result in fewer mutations, so modern humans would display less genetic diversity.
So a person's genetic material "contains the whole history of the population from which you descended," Harpending says.
Earlier studies of genetic material known as mitochondrial DNA and microsatellites supported the notion that a small group of perhaps 5,000 to 20,000 primitive humans migrated from East Africa, spread around the world, a rapidly expanded in population as they replaced other human populations elsewhere in Africa 80,000 years ago, and in Asia 50,000 years ago and Europe about 35,000 years ago.
The new study, however, analyzed mutations called SNPs (single nucleotide polymorphisms) in DNA from the nucleus of human cells studied for the Human Genome Project, the effort to map the entire human genetic blueprint. The analysis indicates there was a bottleneck in the human population - what looks like a sharp reduction in the number of people - when ancestors of modern humans colonized Europe roughly 40,000 years ago.
Researchers are not sure what this means because it conflicts with studies of other kinds of human genetic information, which support the idea that a rapidly expanding African population spread globally and replaced local populations elsewhere.
"If Africans moved out of Africa and then populated the whole world, we would see that in the genetic evidence as an expansion in population size," yet the new study indicated the population shrank instead, Rogers says.
The evidence five years ago indicated migrating Africans did not interbreed with local populations, while the new study indicates they did, Rogers notes, adding that the conflicting genetic data mean "the question is still open."
Harpending says one possible explanation for the new data is that there was a large population of humans who migrated from Africa, yet they kept largely to themselves and mated only to a limited extent with local populations in Europe and elsewhere. Because interbreeding still was uncommon, only a few of the prehistoric European genes were incorporated into the modern human genetic blueprint, giving a false impression that the prehistoric human population collapsed or shrank in size, Harpending says.
Another possibility is that the prehistoric African population was large 100,000 years ago, but only a very small number - perhaps a few dozen - of those Africans migrated to other areas some 80,000 years ago, ultimately replacing local populations. That would explain why the human genetic blueprint could give a false impression that the human population collapsed in size even if it did not. But Harpending believes it is unlikely that such a small number of migrants from Africa could spread globally and ultimately replace other populations.
The original news release can be found here
Posted: Thursday, December 26, 2002
by Vinayak Eswaran
From: Current Anthropology
http://www.journals.uchicago.edu/CA/journal/issues/v43n5/025003/025003.html
A Diffusion Wave out of Africa: The Mechanism of the Modern Human Revolution?
CURRENT ANTHROPOLOGY Volume 43, Number 5, December 2002
EVIDENCE OF HYBRIDIZATION
One interesting aspect of the interaction between modern and archaic humans is that hybridization may have occurred between them. Even proponents of the recent-African-origin model do not deny such hybridization, even if they discount the possibility that it finally led to genetic assimilation. The genetic evidence seems to show few obvious signs of assimilation. That few fossils of clearly hybrid morphology (e.g., Duarte et al. 1999) have been found also suggests that little if any hybridization occurred.
However, this model indicates that the latter inference is not necessarily correct. It shows that it is possible that there was a progressive and complete hybridization of the archaics at the wave front even while there was a low rate of assimilation of archaic neutral genes into the emergent modern population. The model further suggests that, far from being an occasional occurrence of little significance, hybridization, along with natural selection for anatomical modernity, could have been the principal reason for the disappearance of the archaic morphology, thus explaining the apparent "extinction" of archaic humans.
The simulations show that African parentage levels full sharply to zero at the wave front, implying that gene flow is associated only with the wave and does not penetrate beyond. So, if the advantage of anatomical modernity was fundamentally linked to morphology, all signs of hybridization would have appeared only at the wave front; hybrids that attained full modernity would have shown few signs of hybridization, at least in their primary metrics. The simulations show the wave front to be barely 800 km in width, and the region within which clear signs of hybridization would have appeared could have been as narrow as 300 km. Hybrids ahead of this narrow region were close to archaic, while those behind were essentially modern. Thus if the diffusion wave traveled through 3,000 km of Europe between 45,000 and 25,000 years ago, only 10% of the fossils of that period could be expected to have clearly mixed morphologywhich may explain the rarity of obvious hybrids in the fossil record.
Another empirical observation that could be explained by these simulations is the relatively rapid transition that has been recorded to occur at the local level (Mellars 1989). The transition from "progressive" archaic to "essentially" modern could have taken as little as 2,000 to 3,000 years, the time required for the 300-km core of the wave front to pass over a site.
DISAPPEARANCE BY HYBRIDIZATION
The archaics at the wave front would have been progressively hybridized even while the fraction of moderns there increased because of natural selection. An obvious way to demonstrate this hybridization is in terms of the archaic/modern parentage ratiothe ratio of the average African parentage of the local archaics (including not-fully-modern hybrids) to that of moderns in the same area. Because the African parentage of a "pure" archaic is always zero, any nonzero value is due to hybridization. As the maximum African parentage in an area would presumably be found in the moderns, the archaic/modern parentage ratio would normally be less than or equal to unity. The ratio could be used to measure the degree of hybridization in the local archaics, larger values of the ratio indicating a more hybridized population. A ratio of zero would indicate that no hybridization had occurred. A ratio of close to unity would indicate that the local archaics had nearly as many African neutral genes as the local moderns, which could only be due to a complete hybridization of the archaics.
FURTHER EVIDENCE OF ASSIMILATION
Other evidence suggests that assimilation did occur. For example, the unique and ancient ( > 200,000-year-old) polymorphisms that exist in some Asian populations and the high coalescence time obtained in a Melanesian population (Harding et al. 1997, Fullerton et al. 1994) can most easily be explained by archaic assimilation into modern populations. Similar genetic evidence from another locus determining hair and skin pigmentation has suggested assimilation from Eurasian archaic populations (Harding et al. 2000).
There is also fossil evidence in Asia of morphological continuity across the archaic-modern transition (Pope 1992), reinforced by fossil evidence from across the world (Wolpoff et al. 2001). Assimilation could also explain the long-standing continuity of anatomical features such as shovel-shaped incisors in East Asians. Many such traits were presumably neutral. Simulations of the Monte Carlo model (not shown here) indicate that single advantageous alleles would have been even more readily assimilated from archaic populations. Such assimilation from resident archaics could have aided the genetic adaptation of the moderns to local ecological conditions as the wave moved far from Africa.
Posted: Tuesday, December 24, 2002
From: BBC
http://news.bbc.co.uk/2/hi/science/nature/2601635.stm
The theory that we are all descended from early humans who left Africa about 100,000 years ago has again been called into question.
US researchers sifting through data from the human genome project say they have uncovered evidence in support of a rival theory.
Most scientists agree with the idea that our ancestors first spread out of Africa about 1.8 million years ago, conquering other lands.
What happened next is more controversial.
The prevailing theory is that a second exodus from Africa replaced all of the local populations, such as Europe's Neanderthals.
Some anthropologists, however, advocate the so-called multiregional theory, that not all the local populations were replaced.
They think some of these ancient people interbred with African hominids, contributing to the gene pool of modern humans.
The new evidence, published online in the Proceedings of the National Academy of Sciences, is based on an analysis of data from the human genome project - the effort to map the entire human genetic blueprint.
Blood and bones
Researchers led by Henry Harpending, professor of anthropology at Utah University, studied small differences in human DNA known as single nucleotide polymorphisms.
Studying when these mutations appeared gives a window into the ancient past, allowing scientists to trace the rise and fall of early humans in different parts of the world.
"The new data seem to suggest that early human pioneers moving out of Africa starting 80,000 years ago did not completely replace local populations in the rest of the world," he says. "There is instead some sign of interbreeding."
The study suggests that there was a bottleneck in the human population when ancestors of modern humans colonised Europe about 40,000 years ago.
This is a puzzle because earlier human genetic studies have backed the idea that a rapidly expanding African population spread globally and replaced all local populations.
One possibility is that there was limited interbreeding between humans migrating from Africa and local populations in Europe and elsewhere.
'Open question'
Commenting on the research, Professor Chris Stringer, Head of Human Origins at London's Natural History Museum, said that in the last few years the multiregional model of human evolution had been called into question by new data, much of it genetic, showing our species had a recent African origin.
He told BBC News Online: "Arguments now centre on whether we are recently and entirely Out of Africa, or just mainly so.
"Some replacement models, and some genetic data, suggest no interbreeding at all with archaic peoples outside of Africa, while other replacement models allow limited interbreeding with the locals over the short time scale in which they overlapped.
"This new research suggests there could have been some interbreeding, but as the authors recognise, it could have been limited, and whether it happened at all is still an open question."
Posted: Monday, December 23, 2002
Reproduced from BBC, Thursday, 11 May, 2000
http://news.bbc.co.uk/1/hi/sci/tech/745080.stm
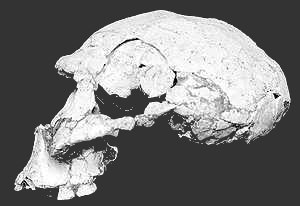 The Remains of what may be the earliest human ancestors to migrate from Africa into Europe have been found in the Republic of Georgia.
The Remains of what may be the earliest human ancestors to migrate from Africa into Europe have been found in the Republic of Georgia.Two skulls, which are probably about 1.7 million years old, were unearthed during an archaeological dig at a medieval castle at Dmanisi.
They were discovered alongside stone tools and the bones of animals. One of the skulls is nearly complete; of the other, only the skullcap has survived.
The fossils suggest that early humans moved out of Africa hundreds of thousands of years earlier than previously thought. They also put a question mark against the assumption that sophisticated stone tool technology was required for migration.
Skeletal characteristics
"This is the first proof for the presence of humans outside Africa at this time," David Lordkipanidze, at the Republic of Georgia State Museum in Tbilisi, told the BBC. "This is most significant."
The age and skeletal characteristics of the Dmanisi skulls link them to the early humans who lived in East Africa at the same time known as Homo ergaster.
This creature was a less developed hominid than Homo erectus that many thought was the first to move out of Africa to populate Asia and Europe.
H. erectus had more sophisticated technology than H. ergaster. He carried a form of advanced tool kit known as the "Acheulean," or "hand-axe" tradition.
But the stone tools found with the Dmanisi fossils are of the less sophisticated "Oldowan" or "pebble-chopper" type that preceded Acheulean technology. More than 1,000 such artefacts were recovered.
The Georgian, German, French and American scientists involved in the latest discovery believe this tool evidence overturns the idea that a more sophisticated technology was required to survive the rigours of migration.
They believe anthropologists will have to look for other explanations for the move.
"It could have been for biological reasons," David Lordkipanidze said. "Humans became carnivores and they wanted to expand their range."
Co-researcher Susan Antón of the University of Florida adds: "Basically, the argument that we're making is that during that time in Africa, the savanna is expanding and there is a greater availability of 'protein on the hoof'.
"With the appearance of Homo, we see bigger bodies that require more energy to run, and therefore need these higher quality sources of protein as fuel."
The Dmanisi discoveries are reported in the journal Science.
Human lines
Commenting on the paper, Dr Chris Stringer, of the Natural History Museum in London, an expert on the "Out of Africa" theory of modern human origins, said is doubtful the Dmanisi hominids were our direct ancestors.
"The line that these people gave rise to may have died out as recently as 50,000 years ago in the Far East," he told the BBC. "So it was a long-lived and successful line, but in my view a dead-end.
"We're descended from similar people who stayed in Africa. Africa in a sense kept pumping out migrations and dispersals of people and this included people like the Neanderthals who, equally, it doesn't seem were our ancestors.
"The ancestors of modern humans may have left Africa only in the last 100,000 years."
Posted: Monday, December 23, 2002
Source: Howard Hughes Medical Institute
A New Window To View How Experiences Rewire The Brain
Howard Hughes Medical Institute researchers have developed sophisticated microscopy techniques that permit them to watch how the brains of live mice are rewired as the mice learn to adapt to new experiences.
Their studies show that rewiring of the brain involves the formation and elimination of synapses, the connections between neurons. The technique offers a new way to examine how learning can spur changes in the organization of neuronal connections in the brain.
The researchers, postdoctoral fellow Josh Trachtenberg, graduate student Brian Chen and Karel Svoboda, a Howard Hughes Medical Institute investigator at Cold Spring Harbor Laboratory, published their findings in the December 19/26, 2002,issue of the journal Nature.
According to Svoboda, researchers had previously shown that the adult brain has a capacity to reorganize in response to new experience. However, it is not clear how this reorganization might occur. Svoboda and his colleagues wanted to see whether learning could induce restructuring of the neural circuitry in the brain that could not be picked up with conventional techniques.
To study those kinds of changes in a living animal, Svoboda and his colleagues started with transgenic mice that were engineered to produce green fluorescent protein within neurons in a portion of the brain that processes tactile sensory inputs from the whiskers. To observe changes in these neurons at high resolution, the scientists constructed a 2-photon laser scanning microscope. This microscope uses an infrared laser to excite green fluorescent protein in neurons, deep in the brain, through a tiny glass window installed in a portion of the mouse's skull.
"Since we had this great tool to look at the brain at unprecedented resolution we did not know what to expect and we began with no preconceived notions of what we might see in these animals," said Svoboda. "Our first observations of the large-scale structure of neurons, their axons and dendrites, revealed that they were remarkably stable over a month." Dendrites and axons are highly branched structures, where dendrites are the input side of neurons and axons the output side.
"However, when we zoomed in closer, we found that some spines on dendrites appeared and disappeared from day to day," said Svoboda. These spines stipple the surface of dendrites, like twigs from a branch, and form the receiving ends of synapses, which are the junctions between neurons where neurotransmitters are released.
"This finding was quite unexpected, because the traditional view of neural development has been that when animals mature, the formation of synapses ceases, which is indicated by stable synaptic densities," said Svoboda. "However, the flaw in this view has been that a stable density only indicates a balanced rate of birth and death of synapses. It doesn't imply the absence of the formation of new synapses, but it was often interpreted that way."
In their experiments, Svoboda and his colleagues observed that about twenty percent of spines disappeared from one day to the next, offset by the formation of new spines.
"While we were surprised at the rate of turnover of some spines, we were also surprised at the incredible stability of other spines," said Svoboda. The spines appeared to fall into different classes. And while there were those that turned over rapidly, other spines, typically the larger ones, persisted for months.
To test whether the new spines were actually forming synapses, the researchers used electron microscopy to analyze in brain slices the same regions that they had studied in the living animals. Those studies revealed that the sprouting spines had indeed formed synapses.
The researchers also explored whether sensory experiences could affect the turnover of spines. In this set of experiments, they trimmed individual whiskers from the mice, forcing them to experience their environment with a subset of whiskers. This manipulation expands the representation of the intact whiskers at the expense of trimmed whiskers. There was a dramatic effect on spine turn-over.
"We found in these animals that there was a pronounced increase in the rate of birth and death of these synapses, as evidenced by increased turnover of spines," said Svoboda. "This finding indicates that there's a pronounced rewiring of the synaptic circuitry, with the formation of new synapses and the elimination of other synapses," he said.
In an accompanying article published in Nature, researchers led by Wen-Biao Gan of the New York School of Medicine found almost no turnover of spines in a region of the visual cortex they studied in mice. Although the results of the experiments would appear to be contradictory, Svoboda said that is not necessarily the correct conclusion. Svoboda said that the visual cortex in adult animals might exhibit far less spine turnover than the tactile sensory region studied by his group. Also, he said, if the animals in the experiments by Gan and his colleagues lived in a visually impoverished environment, experience-dependent synaptic plasticity might not be as evident.
Svoboda said that his team's results suggest that a "sample and hold" model may operate to drive the plasticity of the adult brain. "We believe that the high turnover that we see might play an important role in neural plasticity, in that the sprouting spines reach out to probe different presynaptic partners on neighboring neurons" said Svoboda. "If a given connection is favorable -- that is, reflecting a desirable kind of brain rewiring -- then these synapses are stabilized and become more permanent. But most of these synapses are not going in the right direction, and they are retracted."
The finding that structural plasticity in the adult brain is limited to synapses and spines could help explain the phenomenon of "critical periods," said Svoboda. As an animal matures, there are certain critical periods early in development during which brain plasticity is highly active. By the time the animal reaches adulthood, plasticity is much reduced.
"It may be that in adulthood, since the large-scale structure of neurons does not change, the brain has become essentially an entangled mesh of neuronal processes. Axons and dendrites are stuck with each other as neighbors for life," said Svoboda. "Each neuron thus may have a limited number of permanent neighbors, and any further rewiring with experience is limited to changes in the spines that connect those neurons."
In further studies, Svoboda and his colleagues plan to explore how brain circuitry changes on a larger-scale, by observing mice engineered to express different fluorescent proteins in different populations of neurons.
The original news release can be found here
Posted: Friday, December 20, 2002
Author: Nicholas Wade
Source: The New York Times
http://www.nytimes.com/2002/12/20/health/20GENE.html
Scientists studying the DNA of 52 human groups from around the world have concluded that people belong to five principal groups corresponding to the major geographical regions of the world: Africa, Europe, Asia, Melanesia and the Americas.
The study, based on scans of the whole human genome, is the most thorough to look for patterns corresponding to major geographical regions. These regions broadly correspond with popular notions of race, the researchers said in interviews.
The researchers did not analyze genes but rather short segments of DNA known as markers, similar to those used in DNA fingerprinting tests, that have no apparent function in the body.
"What this study says is that if you look at enough markers you can identify the geographic region a person comes from," said Dr. Kenneth Kidd of Yale University, an author of the report.
The issue of race and ethnicity has forced itself to biomedical researchers' attention because human populations have different patterns of disease, and advances in decoding DNA have made it possible to try and correlate disease with genetics.
The study, published today in Science, finds that "self-reported population ancestry likely provides a suitable proxy for genetic ancestry." In other words, someone saying he is of European ancestry will have genetic similarities to other Europeans.
Using self-reported ancestry "is less expensive and less intrusive" said Dr. Marcus Feldman of Stanford University, the senior author of the study. Rather than analyzing a person's DNA, a doctor could simply ask his race or continent of origin and gain useful information about their genetic make-up.
Several scientific journal editors have said references to race should be avoided. But a leading population geneticist, Dr. Neil Risch of Stanford University, argued recently that race was a valid area of medical research because it reflects the genetic differences that arose on each continent after the ancestral human population dispersed from its African homeland.
"Neil's article was theoretical and this is the data that backs up what he said," Dr. Feldman said.
The new result is based on blood samples gathered from around the world as part of the Human Genome Diversity Project, though on a much less ambitious scale than originally intended. Dr. Feldman and his colleagues analyzed the DNA of more than 1,000 people at some 400 markers. Because the sites have no particular function, they are free to change or mutate without harming the individual, and can become quite different over the generations.
The Science authors concluded that 95 percent of the genetic variations in the human genome is found in people all over the world, as might be expected for a small ancestral population that dispersed perhaps as recently as 50,000 years ago.
But as the first human populations started reproducing independently from one another, each started to develop its own pattern of genetic differences. The five major continental groups now differ to a small degree, the Science article says, as judged by the markers. The DNA in the genes is subject to different pressures, like those of natural selection.
Similar divisions of the world's population have been implied by earlier studies based on the Y chromosome, carried by males, and on mitochondrial DNA, bequeathed through the female line. But both elements constitute a tiny fraction of the human genome and it was not clear how well they might represent the behavior of the rest of the genome.
Despite the large shared pool of genetic variation, the small number of differences allows the separate genetic history of each major group to be traced. Even though this split broadly corresponds with popular notions of race, the authors of Science article avoid using the word, referring to the genetic patterning they have found with words like "population structure" and "self-reported population ancestry."
But Dr. Feldman said the finding essentially confirmed the popular conception of race. He said precautions should be taken to make sure the new data coming out of genetic studies were not abused.
"We need to get a team of ethicists and anthropologists and some physicians together to address what the consequences of the next phase of genetic analysis is going to be," he said.
Some diseases are much commoner among some ethnic groups than others. Sickle cell anemia is common among Africans, while hemochromatosis, an iron metabolism disorder, occurs in 7.5 percent of Swedes. It can therefore be useful for a doctor to consider a patient's race in diagnosing disease. Researchers seeking the genetic variants that cause such diseases must take race into account because a mixed population may confound their studies.
The new medical interest in race and genetics has left many sociologists and anthropologists beating a different drum in their assertions that race is a cultural idea, not a biological one. The American Sociological Association, for instance, said in a recent statement that "race is a social construct" and warned of the "danger of contributing to the popular conception of race as biological."
Dr. Alan Goodman, a physical anthropologist at Hampshire College and an adviser to the association, said, "there is no biological basis for race." The clusters shown in the Science article were driven by geography, not race, he said.
But Dr. Troy Duster, a sociologist at New York University and chairman of the committee that wrote the sociologists' statement on race, said it was meant to talk about the sociological implications of classifying people by race and was not intended to discuss the genetics.
"Sociologists don't have the competence to go there," he said.
Posted: Friday, December 20, 2002
Source: Space Telescope Science Institute
New detailed images from NASA's Hubble Space Telescope show a "late-blooming" galaxy, a small, distorted system of gas and stars that still appears to be in the process of development, even though most of its galactic cousins are believed to have started forming billions of years ago.
Evidence of the galaxy's youthfulness can be seen in the burst of newborn stars and its disturbed shape. This evidence indicates that the galaxy, called POX 186, formed when two smaller clumps of gas and stars collided less than 100 million years ago (a relatively recent event in the universe's 13-billion-year history), triggering more star formation. Most large galaxies, such as our Milky Way, are thought to have formed the bulk of their stars billions of years ago.
The Hubble images of POX 186 support theories that all galaxies originally formed through the assembly of smaller "building blocks" of gas and stars. These galactic building blocks formed shortly after the Big Bang, the event that created the universe. Astronomers Michael Corbin of the Space Telescope Science Institute in Baltimore, Md., and William Vacca of the Max-Planck Institute for Extraterrestrial Physics in Garching, Germany, used the telescope's Wide Field and Planetary Camera 2 to study POX 186 in March and June 2000. Their results will appear in the Dec. 20 issue of the Astrophysical Journal.
"This is a surprising find," Corbin says. "We didn't expect to see any galaxies forming in the nearby universe. POX 186 lies only about 68 million light-years away, which means that it is relatively close to us in both space and time."
Adds Vacca: "POX 186 may be giving us a glimpse of the early stages of the formation process of all galaxies."
POX 186 is a member of a class of galaxies called blue compact dwarfs because of its small size and its collection of hot blue stars. [The term "POX" is derived from the French "prism objectif," or objective prism, a device that astronomers place in front of a telescope to photograph spectra of all objects in its field of view.] POX 186 was discovered 20 years ago, but ground-based telescopes resolved few details of the galaxy's structure because it is so tiny. To probe the galaxy's complex structure, astronomers used the sharp vision of the Hubble telescope. The Hubble pictures reveal that the system is puny by galaxy standards, measuring only about 900 light-years across, and containing just 10 million stars. By contrast, our Milky Way is about 100,000 light-years across and contains more than 100 billion stars.
So why did POX 186 lag behind its larger galactic cousins in forming? Corbin and Vacca find that the young system sits in a region of comparatively empty space known as a void. Its closest galactic neighbors are about 30 million light-years away. The two small clumps of gas and stars that are merging to form POX 186 would have taken longer to be drawn together by gravity than similar clumps in denser regions of space. The Hubble data don't reveal the ages of the stars in the clumps. Corbin, however, suspects that the oldest stars may be about 1 billion years old, which is young on the cosmic time scale.
The youthful galaxy's puny size may support a recent theory of galaxy formation known as "downsizing," which proposes that the least massive galaxies in the universe are the last to form. In clear contrast to POX 186, the most massive galaxies in the universe, known as giant ellipticals, have a generally spherical structure with few or no young stars, indicating that they formed many billions of years in the past. To actually see the formation process of stars in such large galaxies, astronomers are awaiting the deployment of Hubble's successor, the James Webb Space Telescope. This telescope is designed, in part, to study faint objects whose light left them early in the 13-billion-year history of the universe.
Although the POX 186 results are tantalizing, Corbin and Vacca realize that one galaxy is not enough evidence to support the idea that galaxy formation is an ongoing process. They are proposing to use Hubble to study nine other blue compact dwarfs for similar evidence of recent formation.
The original news release can be found here
Posted: Monday, December 16, 2002
Source: National Science Foundation
Scientists Use South Pole Telescope To Produce The Most Detailed Images Of The Early Universe
Using a powerful new instrument at the South Pole, a team of cosmologists has produced the most detailed images of the early Universe ever recorded. The research team, which was funded by the National Science Foundation (NSF), has made public their measurements of subtle temperature differences in the Cosmic Microwave Background (CMB) radiation.
The CMB is the remnant radiation that escaped from the rapidly cooling Universe about 400,000 years after the Big Bang. Images of the CMB provide researchers with a snapshot of the Universe in its infancy, and can be used to place strong constraints on its constituents and structure. The new results provide additional evidence to support the currently favored model of the Universe in which 30 percent of all energy is a strange form of dark matter that doesn't interact with light and 65 percent is in an even stranger form of dark energy that appears to be causing the expansion of the Universe to accelerate. Only the remaining five percent of the energy in the Universe takes the form of familiar matter like that which makes up planets and stars. The researchers developed a sensitive new instrument, the Arcminute Cosmology Bolometer Array Receiver (ACBAR), to produce high-resolution images of the CMB. ACBAR's detailed images reveal the seeds that grew to form the largest structures seen in the Universe today. These results add to the description of the early Universe provided by several previous ground-, balloon- and space-based experiments. Previous to the ACBAR results, the most sensitive, fine angular scale CMB measurements were produced by the NSF-funded Cosmic Background Investigator (CBI) experiment observing from a mountaintop in Chile.
William Holzapfel, of the University of California at Berkeley and ACBAR co-principal investigator, said it is significant that the new ACBAR results agree with those published by the CBI team despite the very different instruments, observing strategies, analysis techniques, and sources of foreground emission for the two experiments. He added that the new data provide a more rigorous test of the consistency of the new ACBAR results with theoretical predictions.
"It is amazing how precisely our theories can explain the behavior of the Universe when we know so little about the dark matter and dark energy that comprise 95 percent of it," said Holzapfel.
The dark energy inferred from the ACBAR observations may be responsible for the accelerating expansion of the Universe. "It is compelling that we find, in the ancient history of the Universe, evidence for the same dark energy that supernova observations find more recently," said Jeffrey Peterson of Carnegie Mellon University.
The construction of the ACBAR instrument and observations at the South Pole were carried out by a team of researchers from the University of California, Berkeley, Case Western Reserve University, Carnegie Mellon University, the California Institute of Technology, Jet Propulsion Laboratory (JPL), and Cardiff University in the United Kingdom. Principle investigators Holzapfel and John Ruhl at Case Western led the effort, which built and deployed the instrument in only two years.
ACBAR is specifically designed to take advantage of the unique capabilities of the 2.1-meter Viper telescope, built primarily by Jeff Peterson and collaborators at Carnegie Mellon and installed by NSF and its South Pole Station in Antarctica. The receiver is an array of 16 detectors built by Cal Tech and the JPL that create images of the sky in 3-millimeter wavelength bands near the peak in the brightness of the CMB. In order to reach the maximum possible sensitivity, the ACBAR detectors are cooled to two-tenths of a degree above absolute zero, or about –273 degrees Celsius (–459 Fahrenheit). ACBAR has just completed its second season of observations at the South Pole. Researcher Mathew Newcomb kept the telescope observing continuously during the six-month-long austral winter, despite temperatures plunging below –73 degrees Celsius (–100 Fahrenheit).
The construction of ACBAR and Viper was funded as part of the NSF Center for Astrophysical Research in Antarctica. The U.S. Antarctic Program provides continuing support for telescope maintenance, observations, and data analysis. NSF's Amundsen-Scott South Pole Station is ideally suited for astronomy, especially observations of the CMB. The station is located at an altitude of approximately 3,000 meters (10,000 feet), atop the Antarctic ice sheet. Water vapor is the principal cause of atmospheric absorption in broad portions of the electromagnetic spectrum from near infrared to microwave wavelengths. The thin atmosphere above the station is extremely cold and contains almost no water vapor. "Our atmosphere may be essential to life on Earth," said Ruhl, "but we'd love to get rid of it. For our observations, the South Pole is as close as you can get to space while having your feet planted firmly on the ground."
Papers describing the ACBAR CMB angular power spectrum and the constraints it places on cosmological parameters have been submitted to the Astrophysical Journal for publication.
For more information and drafts of the submitted papers, see: http://cosmology.berkeley.edu/group/swlh/acbar
The original news release can be found here
Posted: Friday, December 13, 2002
Source: Joint Genome Institute
WALNUT CREEK, CA -- The streamlined genome of Ciona intestinalis, a common sea squirt closely related to vertebrates on the evolutionary tree, is providing new clues about the origins of key vertebrate systems and structures including the human hormone, nervous and immune systems.
In an article for the December 13, 2002 issue of the journal Science, an international consortium of researchers reports on the draft sequencing, assembly, and analysis of the genome of C. intestinalis. The consortium is led by the U.S. Department of Energy's Joint Genome Institute (JGI); the Department of Molecular and Cellular Biology and Center for Integrative Genomics at the University of California, Berkeley; the Department of Zoology at Kyoto University, Japan; and Japan's National Institute of Genetics in Mishima. It includes nearly two dozen other research institutions.
By comparing Ciona's genome with those of the human and other animals, the researchers were able to glean new insights into the evolutionary origins of the human brain, spine, heart, eye, thyroid gland, and nervous and immune systems, as well as a better understanding of chordate and vertebrate development in general.
"We have known for years that despite their humble appearance, sea squirts, or ascidians, have a number of physical characteristics that are similar to vertebrates," said JGI Director Eddy Rubin. "Through a comparison of their genomes, we can now understand how these relationships are reflected at the molecular level, and how these similar systems and gene sets evolved in different ways over 500 million years from a common ancestor."
The sea squirt is a urochordate, or "tunicate," found in shallow ocean waters around the world. The barrel-shaped adult squirt attaches to rocks, piers, boats and the sea bottom, and feeds by siphoning seawater through its body and using a basket-like internal filter to capture plankton and oxygen.
It is as juveniles, however, that ascidians reveal their kinship to humans. One day after an egg is fertilized, it develops into a small tadpole comprised of only about 2,500 cells. The tadpole soon finds a home, settles down, and metamorphoses into its immobile adult form -- but while still in its swimming stage the tadpole has a stiffened rod running the length of its tail called a notochord, the forerunner of our backbone, as well as a primitive nervous system.
Ciona's close relationship to vertebrates, along with its compact genome -- about 160 million base pairs, one-twentieth the size of the human -- and small cell complement make it an ideal model organism for studying chordate development and DNA regulatory networks. The JGI's comparative genomic studies of multiple organisms, which are funded by the Office of Biological and Environmental Research in DOE's Office of Science as part of its Human Genome Program, are helping to determine the functions and regulation of the genes and other DNA regions in the human genome that play a crucial role in human health.
The Ciona sequence analysis revealed that its genome contains about 16,000 genes, about 80 per cent of which are also found in humans and other vertebrates. The total number of Ciona genes, however, is only about half the number in vertebrates.
Daniel Rokhsar, head of the JGI's computational genomics department, said one reason for this "slimmed-down" DNA sequence is the fact that Ciona has single copies of a large number of genes that are present in multiple copies in vertebrates.
"One of two things can happen to these redundant copies," Rokhsar said. "Either they mutate away and disappear, or they evolve to perform other functions. By looking at Ciona's genome, we can see what innovations occurred in the human lineage that enabled us to advance in complexity."
For example, the Ciona analysis revealed a number of vertebrate-like genes, including complement, lectin, and Toll-like receptor genes, that researchers said may play a role in Ciona's primitive "innate" immune system. The scientists found no evidence, however, of the hundreds of key genes, such as immunoglobulin or T cell receptors, involved in the adaptive immune systems found in higher vertebrates.
Ciona is also missing a number of the important Hox genes that determine vertebrate body structure. In some cases only single Hox genes are present, while some appear to be entirely absent. On the other hand, Ciona has light-sensing genes very similar to those of vertebrates, as well as genes involved in the formation of the heart and of cells that accumulate iodine, much like the human thyroid gland. Ciona, however, uses a protein called hemocyanin -- rather than the hemoglobin used by most other animals -- to transport oxygen through its bloodstream, and it lacks the genes to produce steroids and histamines.
The researchers were surprised to find that in some ways, Ciona has more in common with bacteria, fungi and plants than with vertebrates: the genes it uses to produce the fibrous tunic that supports its feeding siphons are similar to the genes involved in metabolizing cellulose.
"Because cellulose is typically produced only by plants and bacteria, its presence in ascidians is a curious lineage-specific evolutionary innovation," the researchers said.
"Just like vertebrates, modern ascidians have been evolving in their own way from our common origins," said Rokhsar. "The Ciona genome reflects the ways they have mobilized many of the same genetic raw materials that we both inherited to serve their own specialized needs."
Mike Levine, professor of genetics and development at UC Berkeley, noted that the Ciona sequence analysis is "entirely consistent" with Darwin's 1871 suggestion in The Descent of Man that ascidians and vertebrates diverged from a common ancestor, with, as Darwin put it, "...one branch retrograding in development and producing the present class of Ascidians, the other rising to the crown and summit of the animal kingdom by giving birth to the Vertebrata."
Ciona was sequenced using the whole-genome shotgun method, which has also been used to sequence the human, pufferfish, mouse, and rat genomes. Earlier this year the JGI convened a Ciona "annotation jamboree" in Walnut Creek, bringing together an international group of experts from the United States, Japan, Italy, France, Scotland, Canada, and Australia to analyze the sea squirt genome and begin disentangling its evolutionary relationship with other animal genomes.
"Bringing the community together was essential," Rokhsar said. "Only by a collaborative effort could we hope to get a coherent sense of the relationship of the Ciona to other organisms, which in turn illuminates the mechanisms of evolution."
In addition to analyzing the Ciona's genome, JGI researchers are also conducting experiments to help identify the function of non-coding sequences of DNA lying between the genes, which regulate gene expression and determine when and where in the organism a given gene is turned on and off. Understanding the complex orchestration of gene networks is crucial to much of contemporary biomedical research.
Results of those studies are expected to be published in early 2003. Meanwhile, Ciona intestinalis sequence information and additional information on the sea squirt can be accessed on JGI's website at http://jgi.doe.gov/ciona.
###
The Joint Genome Institute, established in 1997, is one of the largest and most productive publicly funded human genome sequencing institutes in the world. The JGI was founded by three DOE national laboratories managed by the University of California: Lawrence Berkeley National Laboratory and Lawrence Livermore National Laboratory in California and Los Alamos National Laboratory in New Mexico. In addition to the Ciona project, the JGI has whole genome sequencing programs that include vertebrates, fungi, plants, and bacteria and other single-celled microbes. Funding for the JGI is predominantly from the Office of Biological and Environmental Research in DOE's Office of Science, with additional funding from NIH, NSF, USDA and NASA. Additional information and progress reports on JGI projects, including daily updates of sequence information and assembly statistics, are available at http://www.jgi.doe.gov.
The National Institute of Genetics (NIG) is located in the city of Mishima near Fuji Hakone National Park. It was established in 1949 as the central institute for studies on various aspects of genetics. In 1984, NIG was reorganized as an inter-university research institute to promote collaborative studies. NIG serves as a stock center for various genetic resources and for the DNA Data Bank of Japan, and provides excellent research activities for inter-university collaborations. In 1988, the Graduate University for Advanced Studies was founded, and NIG now undertakes graduate education at the Department of Genetics. NIG has been exploiting the evolving nature of genetics to extend the frontiers of the life sciences.
The original news release can be found here
Posted: Friday, December 13, 2002
Source: Howard Hughes Medical Institute
Researchers Discover Gene That Controls Ability To Learn Fear
Researchers have discovered the first genetic component of a biochemical pathway in the brain that governs the indelible imprinting of fear-related experiences in memory.
The gene identified by researchers at the Howard Hughes Medical Institute at Columbia University encodes a protein that inhibits the action of the fear-learning circuitry in the brain. Understanding how this protein quells fear may lead to the design of new drugs to treat depression, panic and generalized anxiety disorders.
The findings were reported in the December 13, 2002 issue of the journal Cell, by a research team that included Howard Hughes Medical Institute (HHMI) investigators Eric Kandel at Columbia University and Catherine Dulac at Harvard University. Lead author of the paper was Gleb Shumyatsky, a postdoctoral fellow in Kandel's laboratory at Columbia University. Other members of the research team are at the National Institutes of Health and Harvard Medical School.
According to Kandel, earlier studies indicated that a specific signaling pathway controls fear-related learning, which takes place in a region of the brain called the amygdala. "Given these preliminary analyses, we wanted to take a more systematic approach to obtain a genetic perspective on learned fear," said Kandel.
One of the keys to doing these genetic analyses, Kandel said, was the development of a technique for isolating and comparing the genes of individual cells, which was developed at Columbia by Dulac with HHMI investigator Richard Axel. Shumyatsky applied that technique, called differential screening of single-cell cDNA libraries, to mouse cells to compare the genetic activity of cells from a region of the amygdala called the lateral nucleus, with cells from another region of the brain that is not known to be involved in learned fear. The comparison revealed two candidate genes for fear-related learning that are highly expressed in the amygdala.
The researchers decided to focus further study on one of the genes, Grp, which encodes a short protein called gastrin-releasing peptide (GRP), because they found that this protein has an unusual distribution in the brain and is known to serve as a neurotransmitter. Shumyatsky's analysis revealed that the Grp gene was highly enriched in the lateral nucleus, and in other regions of the brain that feed auditory inputs into the amygdala.
"Gleb's finding that this gene was active not only in the lateral nucleus but also in a number of regions that projected into the lateral nucleus was interesting because it suggested that a whole circuit was involved," said Kandel. Shumyatsky next showed that GRP is expressed by excitatory principal neurons and that its receptor, GRPR, is expressed by inhibitory interneurons. The researchers then undertook collaborative studies with co-author Vadim Bolshakov at Harvard Medical School to characterize cells in the amygdala that expressed receptors for GRP. Those studies in mouse brain slices revealed that GRP acts in the amygdala by exciting a population of inhibitory interneurons in the lateral nucleus that provide feedback and inhibit the principal neurons.
The researchers next explored whether eliminating GRP's activity could affect the ability to learn fear by studying a strain of knockout mice that lacked the receptor for GRP in the brain.
In behavioral experiments, they first trained both the knockout mice and normal mice to associate an initially neutral tone with a subsequent unpleasant electric shock. As a result of the training, the mouse learns that the neutral tone now predicts danger. After the training, the researchers compared the degree to which the two strains of mice showed fear when exposed to the same tone alone -- by measuring the duration of a characteristic freezing response that the animals exhibit when fearful.
"When we compared the mouse strains, we saw a powerful enhancement of learned fear in the knockout mice," said Kandel. Also, he said, the knockout mice showed an enhancement in the learning-related cellular process known as long-term potentiation.
"It is interesting that we saw no other disturbances in these mice," he said. "They showed no increased pain sensitivity; nor did they exhibit increased instinctive fear in other behavioral studies. So, their defect seemed to be quite specific for the learned aspect of fear," he said. Tests of instinctive fear included comparing how both normal and knockout mice behaved in mazes that exposed them to anxiety-provoking environments such as open or lighted areas.
"These findings reveal a biological basis for what had only been previously inferred from psychological studies -- that instinctive fear, chronic anxiety, is different from acquired fear," said Kandel.
In additional behavioral studies, the researchers found that the normal and knockout mice did not differ in spatial learning abilities involving the hippocampus, but not the amygdala, thus genetically demonstrating that these two anatomical structures are different in their function.
According to Kandel, further understanding of the fear-learning pathway could have important implications for treating anxiety disorders. "Since GRP acts to dampen fear, it might be possible in principle to develop drugs that activate the peptide, representing a completely new approach to treating anxiety," he said. However, he emphasized, the discovery of the action of the Grp gene is only the beginning of a long research effort to reveal the other genes in the fear-learning pathway.
More broadly, said Kandel, the fear-learning pathway might provide an invaluable animal model for a range of mental illnesses. "Although one would ultimately like to develop mouse models for various mental illnesses such as schizophrenia and depression, this is very hard to do because we know very little about the biological foundations of most forms of mental illness," he said. "However, we do know something about the neuroanatomical substrates of anxiety states, including both chronic fear and acute fear. We know they are centered in the amygdala.
"And while I don't want to overstate the case, in studies of fear learning we could well have an excellent beginning for animal models of a severe mental illness. We already knew quite a lot about the neural pathways in the brain that are involved in fear learning. And now, we have a way to understand the genetic and biochemical mechanisms underlying those pathways."
The original news release can be found here
Posted: Friday, December 13, 2002
Source: Dartmouth College
Melodies In Your Mind: Researchers Map Brain Areas That Process Tunes
HANOVER, N.H. - Researchers at Dartmouth are getting closer to understanding how some melodies have a tendency to stick in your head or why hearing a particular song can bring back a high school dance. They have found and mapped the area in your brain that processes and tracks music. It's a place that's also active during reasoning and memory retrieval.
The study by Petr Janata, Research Assistant Professor at Dartmouth's Center for Cognitive Neuroscience, and his colleagues is reported in the Dec. 13, 2002, issue of Science. Their results indicate that knowledge about the harmonic relationships of music is maintained in the rostromedial prefrontal cortex, which is centrally located, right behind your forehead. This region is connected to, but different from, the temporal lobe, which is involved in more basic sound processing.
"This region in the front of the brain where we mapped musical activity," says Janata, "is important for a number of functions, like assimilating information that is important to one's self, or mediating interactions between emotional and non-emotional information. Our results provide a stronger foundation for explaining the link between music, emotion and the brain."
Using functional magnetic resonance imaging (fMRI) experiments, the researchers asked their eight subjects, who all had some degree of musical experience, to listen to a piece of original music. The eight-minute melody, composed by Jeffrey Birk, Dartmouth class of '02, when he was a student, moves through all 24 major and minor keys. The music was specifically crafted to shift in particular ways between and around the different keys. These relationships between the keys, representative of Western music, create a geometric pattern that is donut shaped, which is called a torus.
"The piece of music moves around on the surface of the torus, so we had to figure out a way to pick out brain areas that were sensitive to the harmonic motion of the melody," explains Janata. "We developed two different tasks for our subjects to perform. We then constructed a statistical model that separated brain activation due to performing the tasks from the activation that arose from the melody moving around on the torus, independent of the tasks. It was a way to find the pure representation of the underlying musical structure in the brain."
The two tasks involved 1.) asking subjects to identify an embedded test tone that would pop out in some keys but blend into other keys, and 2.) asking subjects to detect sounds that were played by a flute-like instrument rather than the clarinet-like instrument that prevailed in the music. As the subjects performed the tasks, the fMRI scanner provided detailed pictures of brain activity. The researchers compared where the activation was on the donut from moment to moment with the fluctuations they recorded in all regions of the brain. Only the rostromedial prefrontal area reliably tracked the fluctuations on the donut in all the subjects, therefore, the researchers concluded, this area maintains a map of the music.
"Music is such a sought-after stimulus," says Janata. "It's not necessary for human survival, yet something inside us craves it. I think this research helps us understand that craving a little bit more."
Not only did the researchers find and map the areas in the brain that track melodies, they also found that the exact mapping varies from session to session in each subject. This suggests that the map is maintained as a changing or dynamic topography. In other words, each time the subject hears the melody, the same neural circuit tracks it slightly differently. This dynamic map may be the key to understanding why a piece of music might elicit a certain behavior one time, like dancing, and something different another time, like smiling when remembering a dance.
Janata adds, "Distributed and dynamic mapping representations have been proposed by other neuroscientists, and, as far as we know, ours is the first paper to provide empirical evidence for this type of organizational principle in humans."
Not only are these results published in the journal Science, the raw data from this study will also be submitted to the fMRI Data Center at Dartmouth College. The fMRI Data Center provides a publicly accessible repository of peer-reviewed fMRI studies and their underlying data. All traces of personal identity information are removed and the image files are converted into a standard format. This provides access to anyone interested in order to develop and evaluate methods, confirm hypotheses, and perform meta-analyses. It also increases the number of cognitive neuroscientists who can examine, consider, analyze and assess the brain imaging data that have already been collected and published.
Janata's co-authors on the paper are Jeffrey Birk, Dartmouth class of '02, John Van Horn, Research Assistant Professor, Center for Cognitive Neuroscience, Dartmouth; Marc Leman, Ghent University, Belgium; Barbara Tillmann, formerly a Research Associate at Dartmouth, now faculty at Centre National de la Recherché Scientifique in Lyon, France; Jamshed Bharucha, formerly Professor of Psychological and Brain Sciences and Dean of the Faculty of Arts and Sciences at Dartmouth, currently Provost at Tufts University, Medford, Mass. This research is part of the Program Project in Cognitive Neuroscience funded by the National Institutes of Health.
The original news release can be found here
Posted: Wednesday, December 11, 2002
Author: Nicholas Wade
Filed: 12/11/2002
Source: The New York Times
http://www.nytimes.com/2002/12/10/science/10ISLA.html
Inhabitants of the Andaman Islands, a remote archipelago east of India, are direct descendants of the first modern humans to have inhabited Asia, geneticists conclude in a new study.
But the islanders lack a distinctive genetic feature found among Australian aborigines, another early group to leave Africa, suggesting they were part of a separate exodus.
The Andaman Islanders are "arguably the most enigmatic people on our planet," a team of geneticists led by Dr. Erika Hagelberg of the University of Oslo write in the journal Current Biology.
Their physical features — short stature, dark skin, peppercorn hair and large buttocks — are characteristic of African Pygmies. "They look like they belong in Africa, but here they are sitting in this island chain in the middle of the Indian Ocean," said Dr. Peter Underhill of Stanford University, a co-author of the new report.
Adding to the puzzle is that their language, according to Joseph Greenberg, who, before his death in 2001, classified the world's languages, belongs to a family that includes those of Tasmania, Papua New Guinea and Melanesia.
Dr. Hagelberg has undertaken the first genetic analysis of the Andamanese with the help of two Indian colleagues who took blood samples — the islands belong to India — and by analyzing hair gathered almost a century ago by a British anthropologist, Alfred Radcliffe-Brown. The islands were isolated from the outside world until the British set up a penal colony there after the Indian mutiny of 1857.
Only four of the dozen tribes that once inhabited the island survive, with a total population of about 500 people. These include the Jarawa, who still live in the forest, and the Onge, who have been settled by the Indian government.
Genetic analysis of mitochondrial DNA, a genetic element passed down only through women, shows that the Onge and Jarawa people belong to a lineage, known as M, that is common throughout Asia, the geneticists say. This establishes them as Asians, not Africans, among whom a different mitochondrial lineage, called L, is dominant.
The geneticists then looked at the Y chromosome, which is passed down only through men and often gives a more detailed picture of genetic history than the mitochondrial DNA. The Onge and Jarawa men turned out to carry a special change or mutation in the DNA of their Y chromosome that is thought to be indicative of the Paleolithic population of Asia, the hunters and gatherers who preceded the first human settlements.
The mutation, known as Marker 174, occurs among ethnic groups at the periphery of Asia who avoided being swamped by the populations that spread after the agricultural revolution that occurred about 8,000 years ago. It is found in many Japanese, in the Tibetans of the Himalayas and among isolated people of Southeast Asia, like the Hmong.
The discovery of Marker 174 among the Andamanese suggests that they too are part of this relict Paleolithic population, descended from the first modern humans to leave Africa.
Dr. Underhill, an expert on the genetic history of the Y chromosome, said the Paleolithic population of Asia might well have looked as African as the Onge and Jarawa do now, and that people with the appearance of present-day Asians might have emerged only later. It is also possible, he said, that their resemblance to African Pygmies is a human adaptation to living in forests that the two populations developed independently.
A finding of particular interest is that the Andamanese do not carry another Y chromosome signature, known as Marker RPS4Y, that is common among Australian aborigines.
This suggests that there were at least two separate emigrations of modern humans from Africa, Dr. Underhill said. Both probably left northeast Africa by boat 40,000 or 50,000 years ago and pushed slowly along the coastlines of the Arabian Peninsula and India. No archaeological record of these epic journeys has been found, perhaps because the world's oceans were 120 meters lower during the last ice age and the evidence of early human passage is under water.
One group of emigrants that acquired the Marker 174 mutation reached Southeast Asia, including the Andaman islands, and then moved inland and north to Japan, in Dr. Underhill's reconstruction. A second group, carrying the Marker RPS4Y, took a different fork in Southeast Asia, continuing south toward Australia.
Posted: Wednesday, December 11, 2002
Source: Los Alamos National Laboratory
LOS ALAMOS, N.M., Dec. 6, 2002 - Reconnection, the merging of magnetic field lines of opposite polarity near the surface of the sun, Earth and some black holes, is believed to be the root cause of many spectacular astronomical events such as solar flares and coronal mass ejections, but the reason for this is not well understood. Researchers at Los Alamos National Laboratory now have a new theory that may explain the instability and advance the understanding of these phenomena.
Theorists Giovanni Lapenta of Los Alamos National Laboratory's Plasma Theory group and Dana Knoll of the Lab's Fluid Dynamics group presented their findings at the American Geophysical Union meeting in San Francisco at the Moscone Convention Center.
The theory is based on a 19th century mathematical observation called Kelvin-Helmholtz instability. "What we are trying to determine is why magnetic field lines loop out from the surface of the sun, reconnect and then fall back," said Lapenta. "And why these systems, which look very stable, are in fact quite unstable."
According to Lapenta, reconnection rates based on resistivity are orders of magnitude too slow to explain observed coronal reconnections. One possible mechanism that provides fast reconnection rates is known as "driven" reconnection-where external forces drive field lines together in a way that is independent of resistivity. Lapenta and Knoll believe that related work focused on magnetic field line reconnection in Earth's magnetopause has shown that the Kelvin-Helmholtz instability can cause compressive actions that push field lines together and drive reconnection. "We propose that the same mechanism at work in the magnetopause could conceivably be at work in the solar corona and elsewhere," said Lapenta.
In this theory, motion on the visible surface of the sun - the photosphere - leads to twisting deformation waves that move through the chromosphere, a layer of solar atmosphere just above the photosphere, growing larger as they move and emerging with a rapid increase of speed through the sun's corona, or outer atmosphere. This rapid change in speed, or velocity shear, injected into the corona can cause magnetic loops to reconnect, according to Lapenta.
"We have conducted a series of simulations and shown that indeed reconnection can be achieved trough local compression driven by Kelvin- Helmholtz and that the reconnection rate is not sensitive to resistivity," said Lapenta.
From this beginning point, Lapenta hopes to study the processes tied to motion on the surface of the sun to better understand why these "velocity shears" occur and how they move away from the sun and lead to CMEs and other solar events, and to apply this knowledge to better understanding the magnetic fields around the earth and the disc-shaped rotating masses, or accretion discs, that form around some black holes.
###
Los Alamos National Laboratory is operated by the University of California for the National Nuclear Security Administration (NNSA) of the U.S. Department of Energy and works in partnership with NNSA's Sandia and Lawrence Livermore national laboratories to support NNSA in its mission.
Los Alamos enhances global security by ensuring the safety and reliability of the U.S. nuclear stockpile, developing technologies to reduce threats from weapons of mass destruction, and solving problems related to energy, environment, infrastructure, health and national security concerns.
Additional news about Los Alamos is available online at http://www.lanl.gov.
The original news release can be found here
Posted: Wednesday, December 4, 2002
Source: Royal Society
Revolutionary New Theory For Origins Of Life On Earth
A totally new and highly controversial theory on the origin of life on earth, is set to cause a storm in the science world and has implications for the existence of life on other planets. Research* by Professor William Martin of the University of Dusseldorf and Dr Michael Russell of the Scottish Environmental Research Centre in Glasgow, claims that living systems originated from inorganic incubators - small compartments in iron sulphide rocks. The new theory radically departs from existing perceptions of how life developed and it will be published in Philosophical Transactions B, a learned journal produced by the Royal Society.
Since the 1930s the accepted theories for the origins of cells and therefore the origin of life, claim that chemical reactions in the earth's most ancient atmosphere produced the building blocks of life - in essence - life first, cells second and the atmosphere playing a role.
Professor Martin and Dr Russell have long had problems with the existing hypotheses of cell evolution and their theory turns traditional views upside down. They claim that cells came first. The first cells were not living cells but inorganic ones made of iron sulphide and were formed not at the earth's surface but in total darkness at the bottom of the oceans. Life, they say, is a chemical consequence of convection currents through the earth's crust and in principle, this could happen on any wet, rocky planet.
Dr Russell says: "As hydrothermal fluid - rich in compounds such as hydrogen, cyanide, sulphides and carbon monoxide - emerged from the earth's crust at the ocean floor, it reacted inside the tiny metal sulphide cavities. They provided the right microenvironment for chemical reactions to take place. That kept the building blocks of life concentrated at the site where they were formed rather than diffusing away into the ocean. The iron sulphide cells, we argue, is where life began."
One of the implications of Martin and Russell's theory is that life on our planet, even on other planets or some large moons in our own solar system, might be much more likely than previously assumed.
The research by Professor Martin and Dr Russell is backed up by another paper The redox protein construction kit: pre-last universal common+ ancestor evolution of energy-conserving enzymes by F. Baymann, E. Lebrun, M. Brugna, B. Schoepp-Cothenet, M.-T. Giudici-Orticoni & W. Nitschke which will be published in the same edition.
###
*On the origins of cells: a hypothesis for the evolutionary transitions from abiotic geochemistry to chemoautotrophic prokaryotes, and from prokaryotes to nucleated cells by Professor William Martin, Institut fuer Botanik III, University of Dusseldorf and Dr Michael Russell, Scottish Environmental Research Centre, Glasgow.
The original news release can be found here.
Posted: Tuesday, December 3, 2002
Source: University Of Washington
Search For Sympathy Uncovers Patterns Of Brain Activity
Neuroscientists trying to tease out the mechanisms underlying the basis of human sympathy have found that such feelings trigger brain activity not only in areas associated with emotion but also in areas associated with performing an action. But, when people act in socially inappropriate ways this activity is replaced by increased activity in regions associated with social conflict.
Understanding the neurophysiology of such basic human characteristics as sympathy is important because some people lack those feelings and may behave in anti-social ways that can be extremely costly to society, said Dr. Jean Decety of the University of Washington. Decety heads the social-cognitive neuroscience laboratory at the UW's Center for Mind, Brain & Learning and is lead author of a new study that appears in a just-published special issue of the journal Neuropsychologia.
In the study, Decety and doctoral student Thierry Chaminade used positron emission tomography (PET) scans to explore what brain systems were activated while people watched videos of actors telling stories that were either sad or neutral in tone. The neutral stories were based on everyday activities such as cooking and shopping. The sad stories described events that could have happened to anyone, such as a drowning accident or the illness of a close relative. The actors were videotaped telling the stories, which lasted one to two minutes, with three different expressions – neutral, happy or sad.
Decety and Chaminade found that, as people watched the videos, different brain regions were activated depending on whether an actor's expressions matched the emotional content of the story.
When the story content and expression were congruent, neural activity increased in emotional processing areas of the brain – the amygdala and the adjacent orbitofrontal cortex and the insula. In addition, increased activation also was noted in what neuroscientists call the "shared representational" network which includes the right inferior parietal cortex and premotor cortex. This network refers to brain areas that are activated when a person has a mental image of performing an action, actually performs that action or observes someone else performing it.
However, these emotional processing areas were suppressed when the story content and expression were mismatched, such as by having a person smile while telling about his mother's death. Instead, activation was centered in the ventromedial prefrontal cortex and superior frontal gyrus, regions that deal with social conflict.
After watching each video clip, the 12 subjects in the study also were asked to rate the storyteller's mood and likability. Not surprisingly the subjects found the storytellers more likable and felt more sympathetic toward them when their emotional expression matched a story's content than when it did not.
"Sympathy is a very basic way in which we are connected to other people," said Decety. "We feel more sympathy if the person we are interacting with is more like us. When people act in strange ways, you feel that person is not like you.
"It is important to note that the emotional processing network of the brain was not activated when the subjects in our study watched what we would consider to be inappropriate social behavior. Knowing how the brain typically functions in people when they are sympathetic will lead to a better understanding of why some individuals lack sympathy."
The research was funded by France's Institut de la Santé et de la Recherche Médicale, the Talaris Research Institute and the Apex Foundation, the family foundation created by Bruce and Jolene McCaw.
The original news release can be found here
Posted: Tuesday, December 3, 2002
BBC: Tests on skulls found in Mexico suggest they are almost 13,000 years old - and shed fresh light on how humans colonised the Americas.
The human skulls are the oldest tested so far from the continent, and their shape is set to inflame further a controversy over native American burial rights.
The skulls were analysed by a scientist from John Moores University in Liverpool, with help from teams in Oxford and Mexico.
They came from a collection of 27 skeletons of early humans kept at the National Museum of Anthropology in Mexico City.
These were originally discovered more than 100 years ago in the area surrounding the city.
The latest radiocarbon dating techniques allow dating to be carried out on tiny quantities of bone, although the process is expensive.
Dr Silvia Gonzalez, who dated the skull, said: "The museum knew that the remains were of significant historical value but they hadn't been scientifically dated.
"I decided to analyse small bone samples from five skeletons using the latest carbon-dating techniques.
"I think everybody was amazed at how old they were."
The earliest human remains tested prior to this had been dated at approximately 12,000 years ago.
Domestic tools dated at 14,500 years have been found in Chile - but with no associated human remains.
The latest dating is not only confirmation that humans were present in the Americas much earlier than 12,000 years, but also that they were not related to early native Americans.
The two oldest skulls were "dolichocephalic" - that is, long and narrow-headed.
Other, more recent skulls were a different shape - short and broad, like those from native American remains.
Asian travellers
This suggests that humans dispersed within Mexico in two distinct waves, and that a race of long and narrow-headed humans may have lived in north America prior to the American Indians.
Traditionally, American Indians were thought to have been the first to arrive on the continent, crossing from Asia on a land bridge.
Dr Gonzalez told BBC News Online: "We believe that the older race may have come from what is now Japan, via the Pacific islands and perhaps the California coast.
"Mexico appears to have been a crossroads for people spreading across the Americas.
"Our next project is to examine remains found in the Baha penninsula of California, and look at their DNA to see if they are related.
"But this discovery, although it is very significant, raises more questions than it solves."
Scientific analysis of early skull finds in the US has often been halted by native American custom which assumes that any ancient remains involve their ancestors and must be handed over.
However, this evidence that another race may have pre-dated native Americans may strengthen legal challenges from researchers to force access to such remains.
Dr Gonzalez said: "My research could have implications for the ancient burial rights of north American Indians."
Dr Gonzalez has now been awarded a grant from the Mexican government and the UK's Natural Environment Research Council to continue her work for three years.
Reproduced from:
http://news.bbc.co.uk/2/hi/science/nature/2538323.stm
For fair use only
Designed and maintained by: Renee, Hendricks, Vashti, Meri & Amon. S.E.L.F.